The cellular environment is highly crowded with most proteins and RNA/DNA forming homomeric and heteromeric complexes.
Although cryo-electron microscopy (cryo-EM) can analyze the high-resolution protein structure, it cannot establish the functional and dynamic relationship of biological large complexes at the atomic level. Computational biology represented by molecular simulation can make up for the shortcomings of traditional biology in time scale, space scale and dynamics, and clearly describes the flexibility and dynamics of biological large complexes.
A research group led by Prof. LI Guohui from the Dalian Institute of Chemical Physics (DICP) of the Chinese Academy of Sciences (CAS) reviewed recent progress in combining multiscale simulations with cryo-electron microscopy (cryo-EM) analysis to understand large complexes.
The combination of these two technologies could establish the relationship between the static structure and biological function of biological large complexes, so as to better observe and understand the dynamic assembly process and regulation mechanism of biological large complexes.
This work was published in Current Opinion in Structural Biology on August 13.
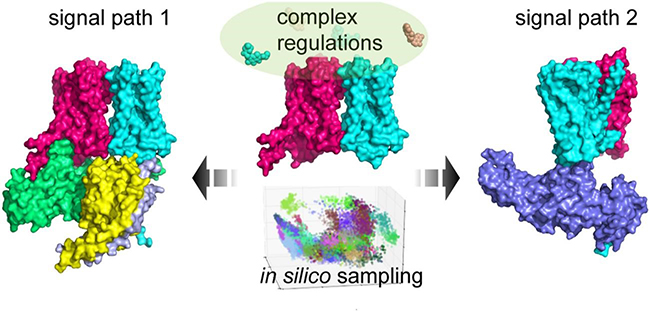
Illustration of the idea to interpret the large complex multi-body regulation in different signaling pathways using single-particle cryo-EM and simulation techniques (Image by LIAO Chenyi and LIU Ye)
The integration of cryo-EM and multiscale molecular modeling can help us to understand the dynamics and function related mechanism in protein-RNA/DNA complexes, protein-protein complexes/assemblies, and membrane protein complexes.
The maturity of single-particle cryo-EM and simulation techniques has brought broad implications in biochemistry molecular biology, cell biology, biophysics, computational biology, chemistry.
The researchers also indicated perspectives to interpret the large complex multi-body regulation in assembly induced function enhancement in conjunction with advanced atomic resolution structural-biology techniques and specialized computing architectures.
This work was supported by the National Natural Science Foundation of China and the Strategic Priority Research Program of Chinese Academy of Sciences. (Text by LIAO Chenyi and LIU Ye)