Diseases are usually resulted from intracellular biological mutation or sharp changes in relative amounts of different components of molecules. And most experimental discovery is focused on the former.
AURKA is critical in cell differentiation, and also plays a vital role in tumor occurrence and development. Past experimental studies have shown that the nuclear AURKA is associated with poor cancer treatment, while, AURKA in cytoplasm does not always induce the formation of cancer, which can indicate that other than general kinase functions, AURKA also plays an unknown and important role. Our experimental collaborator Liu Qiang’s team in Sun Yat-sen's University found a significant increase of AURKA in breast cancer liver cells, and AURKA is formed by interaction with another protein RNPK complex with transcription factor function, activating the MYC promoter, and then control a series of changes related to protein expression in breast cancer. And how is this complex formed, what’s the three-dimensional structure, are essential to answer the above experimental phenomena of molecular mechanisms.
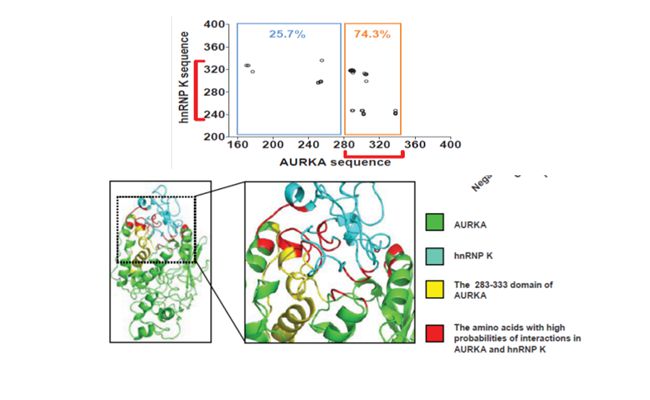
DICP Researchers constructe the Three-dimensional Structure of the Full-length Protein Through a Series of Molecular Modeling Means(Photo by LI Guohui)
By using structural biological means such problems take years to be possible to complete, Molecular modeling is the only alternative in addition to the experiments. Guohui, through a series of molecular modeling means, constructed the three-dimensional structure of the full-length protein. By means of ultra-computing resources and ingenious design of simulation process, he completed the first work of two proteins of molecular recognition process power with 2 microseconds of simulation. Statistical analysis found that the possible modes and their combined interaction sites, the theoretical prediction of binding sites 75% have been experimentally confirmed to be indeed affecting the binding of two proteins, thus providing an important theoretical basis to resolve this complex molecular recognition mechanism. His close work with the experimental workers completely answer microscopic molecular mechanisms of experimental findings. For the diagnosis and treatment of disease, this work provides a new research idea for the next generation of anti-cancer drug development by providing a new design target. Nature Communications 2016 (http://www.nature.com/ncomms/2016/160119/ncomms10180/full/ncomms10180.html).(Text/ Photo by LI Guohui)