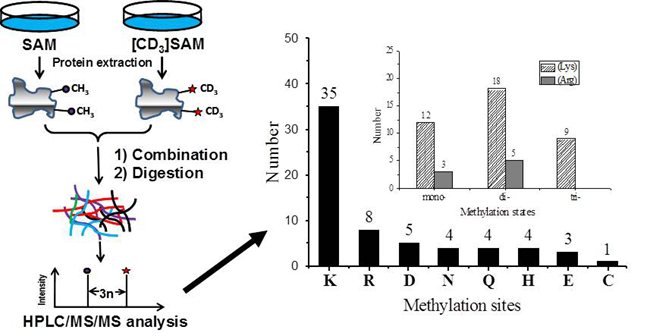
Protein methylation catalyzed by S-adenosyl methionine (SAM)-dependent methyltransferase represents a major posttranslational modification of important biological significance. Because methylation can occur on nitrogen, oxygen and sulfur centres and multiple methylation states exist on the nitrogen centres, methylproteome remains poorly documented. Known methods based on antibody enrichment identified only a part of N-methylation. To address several fundamental questions in protein methylation, Professor Zongbao K. Zhao and Hanfa Zou at Dalian Institute of Chemical Physics, CAS, engineered SAM-auxotrophic Saccharomyces cerevisiae strain, designed the methylation by isotope labelled SAM strategy, and reached a highly-confident yeast methylproteome. Researchers identified 43 methylated proteins, containing 68 methylation events associated with 64 methylation sites. More than 90% of these methylation events were previously unannotated in the Uniprot database. The results indicate, 1) over 2.6% of yeast proteins are methylated, 2) the amino acid residue preference of protein methylation follows the order Lys >> Arg > Asp > Glu ≈ Asp ≈ Gln ≈ His > Cys, 3) N-methylation is largely exclusive in terms of methylation state. The dataset is the most comprehensive methylproteome known-to-date for S. cerevisiae, and should significantly contribute to the field of protein methylation and related research. The study has been published recently at Journal of Proteomics (2015, 114, 226).