Genomic DNA and covalent histone modifications, as benchmarks in epigenetic regulation, have become a hot area of rapid development in the forefront of life sciences, including histone methylation which plays a vital role in regulating transcriptional gene expressions. Mutation of methylation enzyme gene can lead to a variety of abnormal genetic diseases and cancer. Structures of an important class of histone methyltransferase MLL protein complex family have been parsed, illustrating the molecular mechanism of activity regulation. On February 17, 2016, international academic journal Nature published online an article "Structural basis for activity regulation of MLL family methyltransferases" for recent progress of Ming Lei and Yong Chen’s research group in National Center for Protein Science Shanghai, as well as the collaboration work of Guohui Li’s research group in Dalian Institute of Chemical Physics, Chinese Academy of Sciences.
Regulation of MLL Family Methyltransferase Protein Activities via Dynamic Interactions of Multi-protein Complexes (Photo by LI Guohui)
MLL family is a unique group of methylation enzyme of 4-lysine on histone H3. The name was derived from its first discovered member MLL1, because of the Mixed Lineage leukemia caused by its gene rearrangement. MLL family proteins play important roles in gene regulation, cell proliferation, differentiation, growth and development. Dysfunction of MLL caused by mutation can lead to abnormal cell development or function loss, causing various diseases. For example MLL1 gene rearrangements occur in approximately 70-80% of infants suffering from acute lymphoblastic leukemia, and about 5-10% in adult myeloid leukemia (AML). High degree of malignancy of leukemia caused by MLL1 gene rearrangement and poor prognosis leads to a special subtype 11q23 / MLL leukemia, defined by the World Health Organization. On the other hand, MLL2 is highly related to colorectal cancer and breast cancer, and MLL3 mutations are widely observed from patients with gastric cancer, ovarian cancer, colorectal cancer, bile duct cancer, liver cancer, bladder transitional cell carcinoma and other cancers, while MLL4 mutations widespread in Kabuki syndrome, congenital heart disease and other genetic, renal cell carcinoma, non-Hodgkin's lymphoma, prostate cancer, breast cancer, medulloblastoma and other cancers. Because of the important role of MLL family proteins in these diseases, these proteins and macromolecular complexes increasingly become a new scientific target.
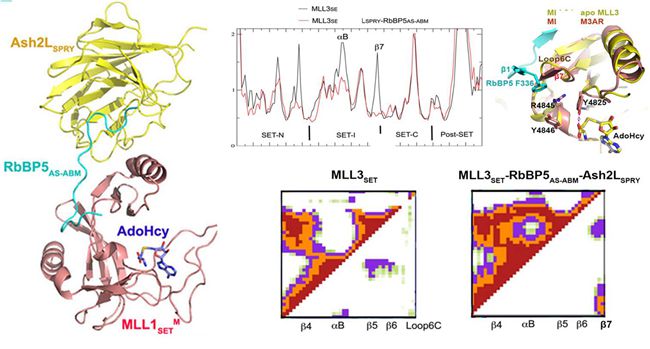
Normal function of MLL proteins relies on a conserved SET domain in its C-terminus performing histone methylation activity, lack of this activity may be an important cause of various diseases. Previous experiments found that different from other methylation enzymes with SET domain, the functionality of MLL proteins requires multiple auxiliary protein including WDR5, RBBP5, ASH2L to effectively complete the methylation process. Due to the lack of atomic-resolution structure, how does the complex effectively implement methylation modification is shrouded in controversy, Whether MLL proteins adopt the same or a different active regulation mechanism remains unknown.
In the present study, the experimenters successfully resolved the MLL proteins in a series of monomeric protein structure and protein complexes, including two MLL family proteins (MLL1 mutants and MLL3) of the SET domain in apo state, in the form of ternary complex with RBBP5-ASH2L, and the crystal structure of active complex with the substrate. They found that in addition to MLL1, activities of other MLL proteins do not depend on WDR5, while merely the complexes with RBBP5-ASH2L will be able to fully activate MLL2/3/4 and SET1A/B proteins. Further research revealed that the RBBP5-ASH2L heterodimer is the smallest unit for binding and activation of MLL proteins, and all of the MLL proteins interact with RBBP5-ASH2L using a conserved mode. Structure comparison revealed that RBBP5-ASH2L did not cause significant MLL SET domain crystal structure changes, but limit the movement of a relatively flexible MLL SET-I module. Molecular dynamics simulations and free energy calculations as well as NMR experiments confirmed that the MLL protein structures in solution are highly dynamic, while adding RBBP5-ASH2L can significantly lead to a fixed and active conformation, which is conducive to the binding of substrate and cofactor, thereby enhancing the MLL methyltransferase activity. This work provided solid structural foundations for further understandings of MLL family in correct complex assemble and activity adjustment. This series of composite structures also provided new targets and clues for the treatment of human MLL subtype leukemia.(Text/Photo by LI Guohui)